Cogent NGS Discovery Software v1.0 User Manual
(Last updated: 01-Oct-2020)
Cogent NGS Discovery Software (referred to as CogentDS in this manual) is bioinformatics software for user-friendly analysis of sequencing data derived from Takara Bio applications, such as the ICELL8 cx Single-Cell system and the ICELL8 Single-Cell system workflows.
I. Before you begin
A. Supported operating systems
The CogentDS is designed to be installed on a user workstation and should work on any system that supports R (see Section I.D). Installation and functionality have been tested and supported for the following OSs:
- Windows 7, Windows 10
- macOS Sierra (10.12) or higher
- Linux CentOS 6.9 or higher
B. Hardware requirements
CogentDS with its dependencies is a lightweight program. It should work on any basic workstation (desktop or laptop) with >2 GB of free disk space and a minimum of 8 GB RAM.
C. User account requirements
By default, administrative privileges are not required to install or run CogentDS. However, if working in an environment where R is installed with IT restrictions, an administrator may need to install the necessary software dependencies (Section I.D) and the Cogent NGS Discovery Software.
D. Additional hardware and software dependencies and recommendations
- Internet connectivity on the computer/server
- R
R is a free, open-source software for statistical computing that provides support across a variety of operating systems. CogentDS is designed to work within an R environment. More information on obtaining and installing R is available in Section III.A.
- RStudio (IDE for R)
RStudio is a free, open-source program that provides graphical user interface (GUI) access to R. More information on obtaining and installing RStudio is available in Section III.C.
- devtools
devtools is a free, open-source R tool that enhances the development and installation of R packages; it's used to install Cogent NGS Discovery Software. More information on obtaining and installing RStudio devtools is available in Section III.D.
- An open network port on the install machine
As the CogentDS interface is accessed through a web GUI, a network port needs to be available on the computer it will be installed on. The port number is selected at random by the Rstudio package, shiny, checking for open ports on the install computer or server until an one is found. For more information about this assignment process, please see https://shiny.rstudio.com/reference/shiny/1.0.1/runApp.html.
If running in an environment where the TCP/IP ports are locked down, please check with your local IT to ensure a port is available on the computer for CogentDS to use.
- Pandoc (optional)
Pandoc is another R utility that is installed natively with RStudio Desktop. For advanced users that wish to forgo RStudio and run R from the command line, the Pandoc Software Package must be downloaded, installed, and placed in the PATH.
E. Required input files
CogentDS requires one of the two following file options as input. The listed files are output from Cogent NGS Analysis Pipeline v1.0 (CogentAP):
CogentDS.analysis.rda, an R-object file. The advantage to this input is that the quality control, clustering, and differential expression modules have been pre-calculated, resulting in faster upload speeds.- Raw gene-count matrix and stats/metadata files. These allow the user to interactively run the data through quality control filtering, clustering, and differential expression. More information on this process can be found in Section IV.D.
II. Software overview

Figure 1. High-level analysis workflow of Cogent NGS Analysis Pipeline (CogentAP) and Cogent NGS Discovery Software (CogentDS).
Figure 1 depicts the high-level workflow of the analysis provided by CogentAP and how its output can be carried over to CogentDS. For more information about Cogent NGS Analysis Pipeline, see the Cogent NGS Analysis Pipeline v1.0 User Manual.
Once CogentDS and required dependencies are installed, analysis can be launched in an interactive RStudio session.
III. Installation and configuration requirements
To obtain Cogent NGS Discovery Software, please visit the ICELL8 software portal at takarabio.com/ICELL8-software.
A. Install R
R and many of the contributed packages are available on the Comprehensive R Archive Network (CRAN). If R is not installed on your system, please download and install R version 3.6.0 or higher from https://www.r-project.org, by first choosing a CRAN mirror of your choice.
For more information on installing R, see the tutorial at datacamp.com.
B. Install platform-specific tools
Installation of CogentDS on Windows or Macintosh workstations requires additional third-party software be installed prior to installing RStudio.
- Windows
On Windows, R requires Rtools to build and install packages from a source file. Download Rtools from https://cran.r-project.org/bin/windows/Rtools/history.html. During installation, ensure that Rtools is included in the system PATH.
For a more information on installing Rtools, see the instructions at rdocumentation.org.
NOTE: Rtools must be installed in a file path with directory names which do not include spaces. (i.e., it cannot be installed in
C:\Program Files\but could be installed inC:\Program\). Installing it in a file path with spaces in the directory names will cause the Cogent NGS Discovery Software installation to fail.If Rtools is installed in such a location on the target computer, please uninstall Rtools and re-install in a folder with a path that conforms to these requirements.
- macOS
R version 3.6.x requires
clang-7.0.0.pkg. This can be downloaded from https://cran.r-project.org/bin/macosx/tools/.
C. Install RStudio
If RStudio is not installed on your system, please download and install the RStudio Desktop (Open Source License) version for your Operating System from rstudio.com.

Figure 2. rstudio.com screenshot of the package to download.
For more information on installing RStudio on Windows, refer to the same tutorial at datacamp.org as in Section III.A for installing R (scroll down towards the bottom of that page).
D. Install devtools
CogentDS requires devtools version 2.1.0 or higher be installed on the computer prior to its installation. Refer to https://cran.r-project.org/web/packages/devtools/readme/README.html.
-
If devtools is already installed on the computer, verify that the version is 2.1.0 or later by running the following command from the Console prompt of RStudio:
packageVersion("devtools")
Figure 3. Example devtools version check in RStudio.
-
To update the devtools version or for a new install, enter the following command into the Console window of RStudio:
install.packages("devtools")
E. (Optional) Install Pandoc
Instructions for downloading and installing Pandoc can be found at pandoc.org.
F. Install Cogent NGS Discovery Software
Once the prerequisites are installed, CogentDS can be installed with the following command:
devtools::install_github("takarabiousa/CogentDS", auth_token = "<AUTHCODE>")
where <AUTHCODE> will be a unique authorization token provided via email (see details below).


Figure 4. Example of the text displayed when the Cogent NGS Discovery Software installation starts.
To obtain the authorization token, please register at the Cogent NGS Discovery Software page at takarabio.com/ICELL8-software.
For first-time users, the installation process may take 10–20 minutes, as many dependencies are automatically downloaded and installed. The installation may also prompt the user to accept downloading and installing certain packages from the source. Answer yes to any such prompts.
G. Upgrading Cogent NGS Discovery Software
The procedure to upgrade CogentDS is to the same as the procedure for doing an installation (Section III.F, above).
It may be that during the upgrade, the script will notice updates to the R dependencies installed along with CogentDS. If that occurs, it is recommended to select whatever the 'All' value is (1 in the Figure 5 example, below).

Figure 5. Example of CogentDS upgrade process detecting newer R dependency packages.
If an error is thrown indicating Rstudio could not remove a prior package installation, please refer to the Cogent NGS Discovery Software notices for one potential fix.
H. Uninstalling Cogent NGS Discovery Software
To uninstall Cogent NGS Discovery Software, run the following command at the Rstudio prompt:
remove.packages("CogentDS")
IV. Launching Cogent NGS Discovery Software
A. Getting started
Once installation is complete, CogentDS can be launched with the following command in an open RStudio session:
CogentDS::launch()
![]()
Entering this command will launch the default browser on your computer and create a new instance of the CogentDS user interface (GUI), running through the localhost of your computer (IP address 127.0.0.1) and a randomly assigned, available TCP/IP port (see Section I.D, for more information about the selection of the TCP/IP port).

Figure 6. Initial screen of CogentDS in the web browser.
B. Upload data
Click [Get Started] to start the process. The Select Input Data window will pop up. Choose the input data type from the dropdown menu, which displays 'CogentDS file' by default.


Figure 7. The Select Input Data browser pop-up window. Default view upon launch and dropdown menu for input options.
NOTE: The Select Input Data menu can also be accessed with the [Upload New Dataset] button from the File Management menu in an established CogentDS session. See Figure 51 in Section IV.F for where to locate the button.
The Select Input Data window allows the user to enter data in one of two ways:
-
Through a
CogentDS.analysis.rdafile, an R object file, which is output from CogentAP.
Figure 8. R object file included in the example data in CogentDS.
-
Through raw gene matrix and stats/metadata files, also output from CogentAP. Refer to Section IV.C for more information about these files.

Figure 9. Data CSV files included in the example data of CogentDS.
The primary difference between the two options is that the CogentDS R file has already been run through the quality control, clustering, and differential expression modules in the Cogent NGS Analysis Pipeline, thus allowing quick visualization of the data.
If a CogentDS R file (Figure 10) is entered, skip to Section IV.E (Discover).


Figure 10. Selecting the example Cogent NGS Analysis Pipeline output CogentDS file or CSV gene matrix and metadata files for input.
Entering the gene matrix and stats files allows the user to exercise more control over parameters by interactively running the data through quality control filtering, clustering, and differential expression analysis described in the rest of this section.
Once the input data source is selected, hit [Submit] to continue.
NOTE: The
Example Datalink in Figures 11 and 15 downloads data from the study "Massively parallel nanowell-based single-cell gene expression profiling" (Goldstein et al. 2017).The screenshots in the rest of this section are based on that sample data.

Figure 11. Download prompt for the sample Goldstein et al. 2017 sample data, from the Example Data link of the CogentDS UI.
C. Format of the gene-count matrix and stats/metadata files
-
Gene matrix file
The gene matrix file (sometimes called the gene table or counts matrix) must be in comma-separated values (CSV) format containing columns of unique sample identifiers, with rows of gene names. Each entry in the matrix is an expression value representing the expression of a gene (i) for a sample (j). The expression data should be raw count data.
If imported from CogentAP, the file will be called
analysis_genematrix.csv(from the GUI) oranalyze_genematrix.csvoranalyze_umi_genematrix.csv(from the command-line interface [CLI]). In Figure 9, it is the zippedgenematrix.csv.NOTE: For optimal performance, compress (.zip) the gene matrix file before uploading.

Figure 12. Example of a gene matrix file.
-
Stats file
The stats file must be in CSV format with one column containing the unique sample identifiers used in the gene-count matrix and any number of subsequent columns with metadata for each sample (i.e., cell type, gene counts, read depth, mitochondrial %, etc.).
If imported from CogentAP, the file will be called
analysis_stats.csv(from the GUI) oranalyze_stats.csv(from the CLI). In Figure 9, it ismetadata.csv.The full list of possible columns in this file accepted by CogentDS is described in the Appendix.

Figure 13. Example Stats file output.
D. Run Quality Control and Clustering modules on raw datasets
Entering the raw gene-count matrix and stats/metadata files prompts the user to enter options for the Quality Control module.
-
The first screen is for QC filter options. Select how to filter non-informative cells and genes from the gene-count matrix. Click the [Next: Norm/Log] button when finished.

Figure 14. Default Quality Control menu for QC filter options.
-
The next screen is for normalization and transformation options.

Figure 15. Default Quality Control menu, for normalization and transformation options.
-
"Normalize gene matrix?"
The available normalization methods include Counts Per Million (CPM), Transcripts Per Kilobase Million (TPM), and Reads Per Kilobase Million (RPKM).

Figure 16. Expanded "Normalization method" drop-down menu.
To normalize by 'median cell coverage', select 'CPM' from the "Normalization method" drop-down menu and type 'median' into the "Normalization factor" input box (Figure 17).

Figure 17. Parameters to normalize by median cell coverage.
-
"Log transform gene matrix?"
To log transform the data, the available options are natural log, Base 2, and Base 10.

Figure 18. Expanded "Log base" drop-down menu.
-
-
When all desired parameters are populated, click [Next: Clustering].
-
A window will pop up prompting you to enter options for the Cluster Analysis module. By default, CogentDS performs clustering analysis based on the 2,000 most variable genes. These genes are used as input for a Principal Component Analysis, and the top Principal Components are selected for further reduction and visualization using either the Uniform Manifold Approximation and Projection (UMAP, recommended) or t-Distributed Stochastic Neighbor Embedding (t-SNE) method.

Figure 19. Cluster Analysis menu.
-
Input your parameters and click [Launch Analysis]. Data transformation will begin, and a status pop-up similar to those in Figure 20 will display on the bottom right-hand corner of the browser window.


Figure 20. Two of the stages of the status pop-up while the cluster analysis is running.
-
After running the cluster analysis, the plot will be rendered in the center of the window, with menus in the right and left sidebars, plus a QC table below the plot.
E. Discover
The baseline plot displays the results of graph-based clustering; however, this can be modified with the User Controls and Formatting menu options in the sidebar panel to the left of the screen.

Figure 21. Baseline UMAP analysis plot.
-
User Controls
-
To highlight cells by cell type, go to the User Controls accordion menu in the left sidebar and select an option from the "Select Group(s)" drop-down box. This field is pre-populated with all column headers from the metadata file. Selecting different metadata features allows the user to highlight the cells by any desired method.


Figure 22. Example "Select Group(s)" drop-down menu and the resulting UMAP analysis plot with cells highlighted by cell sample type.
-
Another method to highlight cells is by expression levels for genes. Entering one or multiple genes into the "Select Gene(s)" field plots the average expression across the panel for each cell and renders the expression into the plot. In Figure 23, the plot highlights ENSG00000261857, a single marker for the A375 cell type.


Figure 23. Example "Select Gene(s)""drop-down menu and the resulting UMAP analysis plot with cells highlighted for gene ENSG00000261857.
-
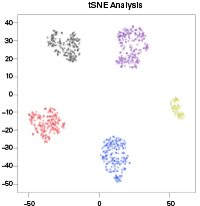
If the user chooses the t-SNE method, the "Perplexity" parameter can also be configured from within the Clustering menu in the right sidebar (see Figure 24). A feature of the t-SNE calculation that broadly serves as an estimate of the cluster size(s) within the data, high perplexity parameters will define large, global structures within the dataset, while smaller perplexities will identify small, local structures.
NOTE: Perplexity defaults have been optimized for general-use cases of the ICELL8 cx Single-Cell and ICELL8 Single-Cell Systems. These values are different from the standard defaults in the Rtsne package and may need to be reoptimized for unique applications.
For more information, please refer to https://github.com/jkrijthe/Rtsne, https://distill.pub/2016/misread-tsne, and https://cran.r-project.org/web/packages/Rtsne/Rtsne.pdf.


Figure 24. t-SNE analysis plot with cells clustered by high perplexity (100, top) and low perplexity (5, bottom).
-
-
Formatting
In the Formatting tab in the left sidebar panel, the marker size and transparency can be changed. These can be used to visualize the data to the user's preferences but are also useful for identifying individual cells within larger clusters.


a. b. 

c. d. Figure 25. Illustration of different marker sizes and transparency selections in the Formatting menu applied to the same data plot. (Top) (a) larger and (b) smaller marker size. (Bottom) (c) less and (d) more transparency.
-
Floating menu
To the right of the chart, there is a menu of icons that only displays when hovered over with the mouse cursor.

Figure 26. Location of the floating menu icons to the right of the UMAP chart.
-
Pan and Reset axes


Figure 27. Identification of the Pan and Reset axes icons in the floating menu.
-
The [Pan] function can be used to move the scatter plot within the frame of the chart axes, changing not only which plots are visible, but also the range values on the X- and Y-axes.

Figure 28. Example after using the Pan function to move the plots down the page, increasing the values of the Y-axis compared to the default in Figure 26.
-
[Reset axes] will return the plot to the default view (Figure 26) after using the [Pan], [Zoom in], and/or [Zoom out] functions.
-
-
Zoom in and Zoom out


Figure 29. Identification of the Zoom in and Zoom out icons in the floating menu.
The [Zoom in] and [Zoom out] buttons can be used to either enlarge or shrink the plots within the chart, decreasing or increasing the scale of the axes (respectively).
-
Lasso Select

Figure 30. Identification of the Lasso Select icon in the floating menu.
The [Lasso Select] feature can be used to select, group, and label cells in a custom manner.
-
Click the [Lasso Select] icon.
-
Click in the plot area and, while holding the mouse button down, use the mouse cursor to draw around the cells of interest. The line will automatically adjust its shape based on the movement of the mouse cursor.

Figure 31. Lassoing a cell cluster of interest.
-
Stop pressing on the mouse button, and the Custom Selection window will pop up.

Figure 32. Default Custom Selection pop-up window.
-
Enter label for selected cells: (Optional) Type in text that will identify the cluster in the legend on the right-hand side of the chart.

Figure 33. Typing in a custom label name for the lassoed cells on the chart.
-
Select color: (Optional) Click on the color bar to expand out to a color selector gradient. Macro changes can be made on the vertical rainbow bar, while finer gradients can be selected by moving the dot around the larger square of color shades on the left.


Figure 34. Examples of two different color selections using the macro and finer gradients fields.
-
-
Once the options are selected, hit the [Set custom label] button to apply them. To quit without applying the customization, press the [Dismiss] button.
-
If the customizations are set, the Custom Selection pop-up will disappear, and the chart will reflect the changes made.

Figure 35. Illustrating the application of the customizations from iii.1, above.
-
Repeat this process, if desired, for other clusters (as shown in the figure below). Or to reset back to the default, click the [Clear custom selections] button, located in the User Controls menu in the left sidebar.


Figure 36. Multiple custom labeled and tinted clusters and the [Clear custom selections] button.
-
-
-
QC table
The Quality Control table under the main figure provides data on QC-filtering and experimental overview statistics.

Figure 37. Example QC table, located in the center frame, below the main analysis figure.
-
Samples: the total number of cells/samples identified for the current analysis
-
Graph-based Clusters Post-QC : the number of cells/samples that remain after QC-filtering
The remaining rows contain experimental overview statistics:
-
Barcoded Reads: total number of barcoded reads found for the experiment
-
Barcodes Identified: total number of barcodes identified
-
Reads per Barcode: average reads per barcode
By clicking on the info icon in the first column of the second row (see Figure 38, top), the user can view a detailed overview of the QC methods that were selected for the current analysis (Figure 38, bottom).


Figure 38. Info icon in the first column of the second row of the QC table (top). Provides detailed overview of QC methods currently in use (bottom).
-
-
Gene Discovery menu
The Gene Discovery menu in the upper-right sidebar contains information and user controls to find genes that distinguish individual clusters.

Figure 39. Gene Discovery menu.
The data in this menu is produced from the differential expression (DE) module. The module calculates DE using the Wilcoxon Rank Sum test with a Benjamini-Hochberg correction for multiple comparisons. The test is performed for each gene for each group within the selected track. The within-group expression is tested against the background, skipping genes with either <0.5 natural logFC or expressed in <10% of both groups.
The results are displayed in the Gene Discovery table, with the top 500 most differentially expressed genes displayed for the selected group. Scroll through the table to view genes of interest.
Clicking on a given gene will display the expression on the UMAP/t-SNE plot.

Figure 40. Display of expression of specific gene (left) selected in the Gene Discovery menu (highlighted in blue, right).
-
For a full list of the differentially expressed genes, click the download link at the top right of the table.

Figure 41. Download icon for the full list of differentially expressed genes. The list is downloaded as a
.csvfile. -
For graph-based clusters, K-means clusters, and any custom track selections, an [Update Cluster Label] button will appear below the Gene Discovery table to allow the user to update the selected cluster label.

Figure 42. Button for updating/customizing cluster labels.
This may be useful for updating generic names such as 'Cluster 1' to informative names, once a cell type has been classified. For example, a cluster (e.g., 'Cluster 1') that has high expression of the gene MS4A1 may be identified as a B-cell cluster. Click to update the generic 'Cluster 1' to the desired label.


Figure 43. Example of 'Cluster 1' label updated to 'B-cell cluster'. This change is reflected both in the plot (left) and Gene Discovery table (right).
-
-
Clustering menu
The Clustering menu contains information and user controls on both dimensionality reduction and cluster identification approaches.

Figure 44. Clustering menu in right sidebar. The Clustering menu contains information and user controls on both dimensionality reduction (PCA and UMAP/t-SNE) and cluster identification approaches (graph-based and K-means clustering).
The data in this menu is produced from the clustering module, which takes the top genes/features identified from the quality controlled, log-normalized data and uses them to perform a Principal Component Analysis (PCA). The top components from this analysis are then determined using a heuristic to find the elbow on the screen plot (with a default minimum of 8 principal components).
The top components are used as the input for clustering and visualization (UMAP/t-SNE) and can be manually set by updating the "Input Principal Components" section of the menu.

Figure 45. "Input Principal Components" section of the Clustering menu.
Clicking the [i] (info) button (Figure 46, top) for this field provides more information and plots to visualize the top principal components (PCs; Figure 46, bottom).


Figure 46. Accessing (top) information and plots (bottom) to visualize the top principal components.
Clustering is performed on the top PCs using either a graph-based approach (adapted from Seurat) or K-means clustering. To switch between these methods, make the desired selection from the "Cluster Method" input within the menu.

Figure 47. "Cluster Method" section of the Clustering menu.
Graph-based clustering is the default method used for identifying clusters. The granularity of the clusters can be changed by setting the "Resolution" parameter within the menu. Values closer to 0 will find broader structure in the data, while values closer to 1 and above will reveal finer structure.


Figure 48. Two UMAP images of the same data with different "Resolution" values in the Clustering menu. The two examples illustrate how different values affect the display.
K-means clusters are calculated for values of K = 2–15 by default. The optimal number of clusters from this set is determined using the Average Silhouette Score Method. Users may also select custom values of K to calculate and observe a desired number of clusters.

Figure 49. K-means Cluster menu.
For visualization of this approach, click the [i] (info) button (Figure 50, top) within the menu.


Figure 50. Accessing information (top) and plots (bottom) to visualize the K-means cluster approach.
F. Export the data
After applying any data manipulations from Section IV.E, it can be saved in its edited form.
-
Expand the File Management option in the left sidebar menu and click the [Download] button.

Figure 51. The File Management submenu.
-
Choose the type of file you wish to download from the dropdown menu. Available options are:
-
a high-resolution UMAP/t-SNE plot
-
an updated stats/metadata CSV file
-
a quality-controlled log-normalized gene matrix
-
a comprehensive HTML report of the analysis
-
a CogentDS R-object file to save changes to the analysis and share with colleagues


Figure 52. The Downloads pop-up menu and dropdown list of available file types.
-
-
If desired, enter a custom file prefix.
-
If downloading an HTML report, two additional fields are available (see Figure 53, below):
-
(Optional) Enter a report author.
-
(Required) Select an option for performing correlation analysis. This module is included in the HTML report and must be run. However, analysis can take a long time, so other options are available to either skip the analysis completely or perform the analysis on only the top genes used for clustering.
NOTE: This HTML report is similar to the one generated from the Cogent NGS Analysis Pipeline but is now customized to your specifications made in CogentDS.

Figure 53. The Downloads pop-up menu for HTML reports.
-
Appendix
Columns in Stats file
The tables below document all potential columns that might display in a Cogent NGS Analysis Pipeline Stats file; they also represent the column names accepted for input into Cogent NGS Discovery Software.
Columns that will be present in all Stats files output by CogentAP (input workflow agnostic).
| Column name | Description |
|---|---|
| Barcode | Detected barcodes. This value will usually be the sample name from the well-list or well-list-like file, but there are three exceptions, documented in the table below. |
| Sample | Sample names described in sample description file. |
| Barcoded_Reads | Number of reads after demultiplexing. |
| Trimmed_Reads | Number of remained reads after trimming. |
| Unmapped_Reads | Number of reads not mapped to genome. |
| Mapped_Reads | Number of reads mapped to genome. |
| Multimapped_Reads | Number of reads mapped to multiple genomic locations. |
| Uniquely_Mapped_Reads | Number of reads mapped to one genomic location. These reads are used for counting. |
| Exon_Reads | Number of reads assigned to an exonic region. |
| Ambiguous_Exon_Reads | Number of reads assigned to exonic regions of multiple genes. |
| Intron_Reads | Number of reads assigned to an intronic region. |
| Ambiguous_Intron_Reads | Number of reads assigned to intronic regions of multiple genes. |
| Gene_Reads | Number of reads assigned to a gene region (exon + intron). |
| Intergenic_Reads | Number of reads assigned to an intergenic region. |
| No_of_Genes | Number of detected genes. |
| Mitochondrial_Reads | Number of reads assigned to mitochondrial chromosome. |
| Ribosomal_Reads | Number of reads assigned to a ribosomal gene. |
The "Barcode" column, in addition to the samples named in the well-list or well-list-like file, will also have three additional rows, which are described in the following table.
| Barcode field value | Description |
|---|---|
| Short | Number of reads containing N in barcode or having shorter length than barcode. |
| Unselected | Number of reads having a barcode included in Chip’s description, but not included in sample description file. |
| Undetermined | Number of reads having undetermined barcode. |
Additional columns in the Stats file for 3′ DE analysis with UMIs on ICELL8 system.
The table below lists additional columns that will be present in the Stats file when the input FASTQ files result from the ICELL8 3′ DE for UMI Reagent Kit (Cat # 640005) workflow on the ICELL8 Single-Cell System (Cat # 640000).
| Column name | Description |
| No_of_UMIs | Number of UMI variations detected after demultiplexing. |
| Exon_nUMIs | Number of deduplicated reads assigned to an exonic region. Deduplication is done by UMI. |
| Intron_nUMIs | Number of deduplicated reads assigned to an intronic region. Deduplication is done by UMI. |
| Gene_nUMIs | Number of deduplicated reads assigned to a gene region (exon + intron). Deduplication is done by UMI. |
Additional columns in Stats file for the SMARTer Stranded Total RNA-Seq Kit v3- Pico Input Mammalian protocol.
The table below lists additional columns that will be present in the Stats file when the input FASTQ files result from the SMARTer Stranded Total RNA-Seq Kit v3- Pico Input Mammalian protocol.
| Column name | Description |
| No_of_UMIs | Number of UMI variations detected after demultiplexing. |
| Exon_nUMIs | Number of deduplicated reads assigned to an exonic region. Deduplication is done by UMI. |
| Exon_nUSSs | Number of deduplicated reads assigned to an exonic region. Deduplication is done by Unique Start&Stop Site (USS). |
| Exon_nUMIs_USSs | Number of deduplicated reads assigned to an exonic region. Deduplication is done by both UMI and USS. |
| Intron_nUMIs | Number of deduplicated reads assigned to an intronic region. Deduplication is done by UMI. |
| Intron_nUSSs | Number of deduplicated reads assigned to an intronic region. Deduplication is done by Unique Start&Stop Site (USS). |
| Intron_nUMIs_USSs | Number of deduplicated reads assigned to an intronic region. Deduplication is done by both UMI and USS. |
| Gene_nUMIs | Number of deduplicated reads assigned to a gene region (exon + intron). Deduplication is done by UMI. |
| Gene_nUSSs | Number of deduplicated reads assigned to a gene region (exon + intron). Deduplication is done by Unique Start&Stop Site (USS). |
| Gene_nUMIs_USSs | Number of deduplicated reads assigned to a gene region (exon + intron). Deduplication is done by both UMI and USS. |
| Strand_Specificity | Ratio of reads detected as correct strand after mapping to genome. |
RNA-seq
Cogent NGS Analysis Pipeline
Analyze sequencing data generated by select Takara Bio applications.
Cogent NGS Discovery Software
Visualize sequencing data using the output from the Cogent NGS Analysis Pipeline.
SMART-Seq DE3 Demultiplexer
Demultiplex sequencing data from SMART-Seq mRNA 3′ DE into sorted read data files.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.