Successful CRISPR knockout experiments—here's what to consider before starting (Part I)
Introduction
With CRISPR in your toolbox (almost) anything's possible. However, a successful knockout experiment involves careful design and planning to achieve the highest on-target activity while also minimizing off-target effects. Important factors to consider before getting started include gathering information about your target gene and cell type/organism, sgRNA design and optimization, mode of delivery of the CRISPR/Cas9 machinery, and methods for verifying knockout efficiencies and characterizing edited cell populations.
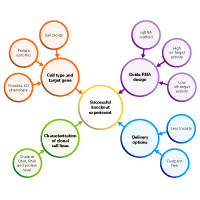
We have a lot of ground to cover, so we have split this post into a two-part blog series. In this first part, we will discuss ideas about how to set up your knockout experiment and the information you need to gather before starting. In the next part, we will discuss the CRISPR-Cas9 delivery options to consider and characterization of the edited cell lines (Figure 1). Follow these simple steps and knockout like a pro!

Figure 1. Key considerations for a successful CRISPR/Cas9 knockout experiment.
Acquiring knowledge about your cell line and target gene
The first step when designing a knockout experiment is to gather information about your target gene and cell line. Factors such as cell ploidy, alternative transcript splicing, and phenotypes associated with the knockout of a given target gene can all contribute to the overall complexity of an experiment.
- Cell ploidy: how many copies of the gene will you need to knock out? When targeting a gene with a high copy number, you may need to select a delivery option that provides a high enough level of Cas9 expression to achieve a complete knockout (i.e., all copies disrupted).
- Sequence and transcripts of your target gene: Ensembl is an online database that will allow you to know the precise sequence (introns and exons) and the transcripts of your gene, which will help you select target sites more effectively (Figure 2).
- Possible phenotypes linked to the knockout: research the literature for previous work related to the function of the target gene to gain insights regarding possible knockout phenotypes, such as lethality, hyperproliferation, etc.

Figure 2. Importance of considering all splice variants of your target gene. During sgRNA design, you should take into account all possible transcripts generated from your target of interest (Panel A). Your sgRNA should target an exon common to all the splice variants of your target gene. In the example above, if you generate an indel in Exon 2, you will still get the expression of Isoform #2 and hence would not get a complete knockout of your gene (Panel B).
Design and optimization of single guide RNAs (sgRNAs)
Once you have identified a suitable target region, design 2–3 sgRNAs with the least predicted off-target effects. Many online tools, such as Chopchop and a few others, will help you predict the off-target activity of your candidate sgRNAs. You could also watch a short video that we created to help you understand how to design sgRNAs.
Considerations for sgRNA design:
- Splice variants: if your target gene encodes splice variants, you need to design sgRNAs that target exons common to all isoforms to achieve a complete knockout. If not, some isoforms would still be expressed in the target cells, providing partial gene function. Another strategy you could follow would be to target relevant functional domains (Shi et al. 2015). See Figure 2 above for an example of how the number of introns and the splice variants of your gene could affect the result of your knockout experiment.
- Using predesigned sgRNAs like the ones in the human CRISPR knockout pooled library (Brunello): these guides have been designed using optimized metrics which combine improved on-target activity predictions with a lower off-target score, by machine learning (GPP Web Portal; Doench et al. 2016).
- Checking for microhomology: one of the likely repair outcomes of double-strand-break repairs could be in-frame deletions. The existence of microhomologies near the cut site can be used to predict the repair outcome via the microhomology-mediated end joining (MMEJ) pathway with more accuracy, and therefore increase the probability of creating knockouts due to specific deletion patterns (Bae et al. 2014). You can predict microhomologies using the Microhomology-Predictor tool.
- Using an optimized sgRNA scaffold: it has been shown that extending the Cas9-binding hairpin and removing four consecutive uracil nucleotides from it improves the knockout efficiency (Dang et al. 2015, Chen et al. 2013). We have incorporated this optimized sequence into all our Guide-it products (Figure 3).
- Sequencing the target site in your target cell line: it is vital to ensure that there are no SNPs in the target site that could affect the activity of your sgRNAs. Extract the genomic DNA from your target cell line, amplify the target gene by PCR, and sequence it to make sure there are no SNPs present before designing your sgRNAs.
- Testing sgRNAs in vitro: not all sgRNAs are born equal; in other words, they do not all have the same activity. Therefore, it is advisable to test sgRNA activity in vitro before proceeding with your experiment. There are in vitro screening kits available that can help you predict the efficiency of candidate sgRNAs before you invest too much time into them.

Figure 3. Optimized sgRNA scaffold improves knockout efficiency. Panel A. Gesicle-mediated knockout with sgRNA containing a traditional scaffold shows a 23% knockout efficiency whereas when sgRNA comprising an optimized scaffold is used, the knockout efficiency improves significantly (59%, Panel B).
Next steps
Knowing your target gene and cell type and designing the best sgRNAs is half the battle. In the next part of this blog series, we will discuss how to choose the right CRISPR/Cas9 delivery method that ensures the best on-target knockout efficiency and minimizes off-target effects so you can get footprint-free editing, as well as methods for postdelivery characterization of your knockout cell populations.
References
Bae, S. et al., Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705–6 (2014).
Chen, B. et al., Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–91 (2013).
Dang, Y. et al., Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Gen. Biol. 16, 280 (2015).
Doench J. G. et al., Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Shi, J. et al., Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015).
Successful CRISPR knockout experiments—here's what to consider before starting (Part II)
The second part of this blog series featuring delivery options and characterization of your knockout cell populations.
Successful knockout experiments part IITakara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.



