Successful CRISPR knockout experiments—here's what to consider before starting (Part II)
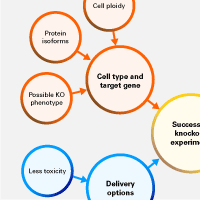
In the first part of this two-part post, we discussed how to gather information about your target gene and cell type/organism, and sgRNA design and optimization. In this post, we will discuss the mode of delivery of the CRISPR/Cas9 machinery, and methods for verifying knockout efficiencies and characterizing edited cell populations (Figure 1).

Figure 1. Key considerations for a successful CRISPR/Cas9 knockout experiment.
Delivery of the CRISPR/Cas9 machinery and analysis of editing
After designing and testing sgRNAs in vitro, you'll need to consider what method of delivery would work best for the cell line with which you are working. You need to pick a method that will work well for your target cell type and minimize cell toxicity caused by the delivery of Cas9 and sgRNA.
Delivery of Cas9 and sgRNA: in our experience, delivery in the form of Cas9-sgRNA ribonucleoprotein complexes (RNPs) either via electroporation or gesicles causes very little toxicity in cells and results in fewer off-target effects compared to plasmid-based delivery. This is largely because RNP delivery results in transient expression of Cas9 and avoids off-target effects triggered by the prolonged presence of plasmids expressing Cas9 under a strong promoter. RNP delivery also avoids dependence on the cellular transcription/translation machinery of the target cells and minimizes undesired integration events resulting in foot-print free genome editing. For cell lines that are hard to transfect or when you work with in vivo models, you could also consider an AAV virus-mediated delivery method.
Checking knockout efficiency: you can check the knockout efficiency in your transfected cell population with a Resolvase assay. You can also characterize the nature of the indels in the overall edited population with an Indel detection kit. Online tools like TIDE or ICE can be employed to perform a fast check of knockout efficiency via amplification of the target site followed by Sanger sequencing of the PCR product. Keep in mind that to accurately perform knockout quantification, high-quality Sanger sequencing traces are essential.
Isolation and expansion of edited cells: once you are confident that the desired knockout has occurred, single cells can be isolated and expanded to obtain isogenic cell lines containing the edit. If you knock out a cell membrane protein, you can isolate the successfully edited population via FACS using an antibody against the extracellular domain of that membrane protein. In other cases, you need to isolate single cells from the whole population and characterize each clonal cell line after its expansion. Single-cell survival and expansion to clonal cell lines can be challenging with certain cell types such as hiPSCs.
Characterization of your knockout clonal cell lines
The last important step in your knockout experiment is to thoroughly characterize the clonal cell lines to make sure that you have achieved a complete knockout of your target gene and that there are no undesired off-target effects.
- DNA level: characterize indels (Indel Identification kit) as well as zygosity using a genotyping kit for edited clones.
- RNA level: check for exon skipping via RT-PCR since the indels created could have affected splicing (Mou et al. 2017, Sharpe and Cooper, 2017).
- Protein level: perform a Western Blot if there is an antibody available (keep in mind that the epitope should be after the targeted site, so truncated proteins will not give false positives) to make sure there is no expression of your target protein.
- Cell-line specific: if the cells are hiPSCs, you should perform karyotyping and pluripotency analysis, and monitor mutations in the p53 gene (Ihry et al. 2018, Haapaniemi et al. 2018).
Summary
A lot goes into designing the best possible CRISPR-knockout experiment, but these tips will help you get started on the right track.
References
Haapaniemi, E. et al., CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Ihry, R. J. et al., p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Mou et al., CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Gen. Biol. 18, 108 (2017).
Sharpe, J. J., and Cooper, T. A. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Gen. Biol. 18, 109 (2017).
Go back to Successful CRISPR knockout experiments—here's what to consider before starting (Part I)
More CRISPR/Cas9 resources
View our video on how to design guide RNAs, or browse all our tools for CRISPR-based genome editing.
Video on guide RNA design Tools for genome editingTakara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.