How to choose the right tools for iPSC-derived disease model development
From infectious diseases to cancer, neurodegenerative diseases, and beyond, countless national and global health initiatives are focused on advancing our understanding of disease mechanisms, developing new treatments, and drug discovery. To achieve this goal, the generation of reliable and translationally relevant disease models is crucial. Traditionally, animal models have been used to simulate human disease, but interspecies genetic differences, however small, can often mean that the findings don't translate.
Recent years have seen the emergence of human induced pluripotent stem cells (hiPSCs) as ideal for disease model research, in no small part due to the accessibility of somatic cells from both healthy and diseased individuals. Gene editing of these cells with CRISPR/Cas9 enables you to perform any number of experiments, including the introduction of putative disease mutations to healthy cells to understand underlying mechanisms, the correction of mutations in unhealthy cells for therapy development, or large-scale screening of novel drugs.
However, creating these models from edited hiPSCs can be extremely challenging, as successful gene editing and concurrent establishment of clonal populations can be both inefficient and time-consuming. With an ever-expanding range of research tools at your fingertips, you will have many options to turn to for either developing disease models yourself, or outsourcing the process so you can instead focus on the research.
With all of the downstream applications that rely on a successful disease model, what criteria should you consider for each stage of development? We've got your list of helpful considerations to ease your decision making.


Culture systems for initial growth
When grown in typical 2D colony cultures, the cell population is not homogenous; cells in the center of the colony are dense while cells at the edges are larger and highly proliferative. Because the overall efficiency of homologous recombination in hiPSCs is low, your chances of successful gene editing are much higher if you have a homogeneous population of cells that are confluent and have the morphology of these "edge cells".
Look for culture systems that set you up for success from the very beginning, enabling you to work from a homogenous starting population.

CRISPR/Cas9 for accurate gene editing
Delivery of CRISPR/Cas9 for gene editing can be done via plasmid or viral-based methods; however, these protocols lead to prolonged expression of Cas9 in the cell, which in turn can increase off-target effects and possible cellular toxicity. To limit its duration in the cell, we strongly recommend high-efficiency electroporation for Cas9-sgRNA complex delivery in the form of a ribonucleoprotein to achieve footprint-free editing—in which cell lines receive only the desired change, without further modification to the genome.
Additional gene editing concerns include sgRNA design and the type of donor template used for homology-directed repair (HDR). Be sure to use an optimized sgRNA scaffold design for enhanced binding to Cas9 and the formation of a more stable complex, which leads to more efficient and successful editing. For knockin experiments, we recommend the use of single-stranded DNA donor templates for HDR, as these have been shown to cause less toxicity and random integration than double-stranded DNA templates.

Screening of edited cell populations
Following gene editing, you will inevitably have a heterogeneous population of cells—some wild type and others with modifications at the target locus. At this point, it is necessary to have a high-throughput, efficient screening method to identify successfully edited clones. Timing is critical, as clonal cell lines will grow at different rates. Identification of edited colonies as early as possible will facilitate their culture and ensure they maintain pluripotency.
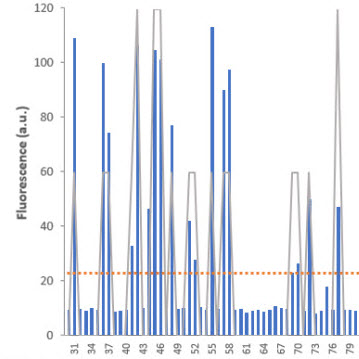
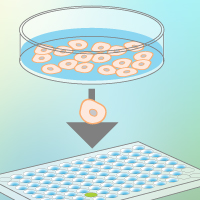
With this in mind, your next step is to isolate single cells from the population and seed them into plates, one cell per well, for accurate screening. Isolation of single cells can be performed via FACS or limiting dilution. Screening methods must be sensitive and accurate, but speed and ease of use are equally important concerns. Opting for systems with fluorescent readouts can make for easy identification of edited clones, and protocols with streamlined workflows and/or high-throughput capabilities can minimize handling, making quick work of your entire cell population.

Culture systems for single-cell cloning and expansion
Once you have isolated and screened your edited hiPSCs, it is time to expand them to generate clonal cell lines with your desired mutation. Culture of individual cells may be challenging, as separation of individual cells with traditional 2D culture methods often leads to a high percentage of cell death or a lack of expansion. Alternatively, if they do expand, the resulting clonal cell line may lose its pluripotency and, therefore, its capacity to differentiate downstream. It is critical to choose a culture system that supports not only the survival of single cells, but also their expansion into clonal cell lines, and maintenance of pluripotency during that expansion.
There is always a risk of mutations occurring when culturing hiPSCs, and this increases the longer they are cultured. It is therefore important to characterize your clonal cell lines by their karyotype. Ideally, even after a high number of passages, your cells would maintain a normal, stable karyotype, making them suitable for downstream applications.
Larger, more robust experiments in clinical and pharmaceutical spaces, including drug screening, greatly benefit from the ability to scale hiPSC culturing. Just like smaller projects, high-throughput applications need homogenous cultures, able to yield high-quality cells that maintain pluripotency and karyotype stability, but on a much larger scale. Variability within cultures becomes a greater concern as the culture size itself increases.

Differentiation systems
Historically, differentiation protocols have been focused squarely on disease-relevant cell types like cardiac, hepatic, pancreatic, and neuronal cells, resulting in the commercial availability of complete solutions for differentiation into these cell types. From among these solutions, pick a robust, proven protocol that gives tightly controlled, stepwise differentiation—that which mimics embryonic development. Following differentiation, you should aim to see a population containing >90% fully differentiated cells that include your desired modification.
When each of these disease model development steps have been successfully executed, your modified, differentiated cells let you perform the rewarding task of screening novel drugs, understanding disease mechanisms, and developing therapeutic treatments. With all the ongoing innovation in the gene editing and stem cell fields and the experiments you're already planning, it is exciting to think about what new techniques will be brought to the bench and what progress will be made in patients' health.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.