Seq-ing the small: metagenomics and metatranscriptomics

In our world of the large, it's easy to overlook our microscopic brethren unless they're directly impacting our lives. A good example is the strains of antibiotic-resistant bacteria (ARB) with a large annual death toll spurring efforts to better understand the microbiome in both clinical and nonclinical settings. But not all microbes are bad: bacteria are excellent at breaking down dry plant matter, such as wood, and the tools that allow them to accomplish this feat can be leveraged to develop more efficient biofuel synthesis methods.
Microbiomes are diverse, complex, and require specialized approaches to investigate. In recent years, the advent of sensitive, high-throughput sequencing methods has allowed for DNA-seq (metagenomics) and RNA-seq (metatranscriptomics) of highly diverse microbial populations. In this blog article, we highlight several groups who showcased the strength of our ThruPLEX DNA-seq and SMARTer Stranded RNA-seq kits for interrogating the microbial world.
Metagenomics and the growing (antibiotic) resistance
Our health is inextricably linked with our environment, and this is particularly true when it comes to microbes. While we take an "out of sight, out of mind" approach to human waste, it influences the microbiome of our surroundings, and these environments can then become a source of transferable antibiotic resistance.
In a report commissioned by the Norwegian Environment Agency, researchers sought to understand the prevalence of antibiotic-resistant bacteria and antibiotic resistance genes (ARGs) in the environment. To accomplish this, they analyzed six wastewater treatment facilities throughout Norway to identify and monitor the prevalence of ARB and ARGs in a nonclinical setting.
Due to the complexity and difficulty of extracting high yields of DNA, ten replicates of untreated wastewater samples from each facility were collected for DNA extraction. Metagenomic libraries were then generated with the ThruPLEX DNA-seq kit. Following sequencing, the researchers identified 110 ARG matches, with 99 corresponding to mex-family efflux pumps. The researchers noted that prior work demonstrated that some of these genes are linked to clinically relevant ARB, and mex-family efflux pumps may also enhance bacterial persistence in hosts and promote pathogenesis (Piddock 2006).
Why is this important?
Metagenomics allows large-scale analyses of microbial population genomics, and this research leveraged this approach to screen environmental samples for ARGs. As the environment can serve as a reservoir for transferable antibiotic resistance, this report underscores the importance of monitoring it in nonclinical settings. However, these analyses can be confounded by the complexity of starting samples, low sample yield, and low-throughput workflows. This report demonstrated the utility of our single-tube ThruPLEX chemistry to generate high-complexity metagenomics libraries from microbial samples taken from the environment.
Metatranscriptomics: turning fibers into fuel
Termites are (in)famous for their ability to eat wood, but their gut microbes are what let them digest its complex carbohydrates. The termite gut, therefore, represents a potential source of novel enzymes for processing lignocellulosic (plant dry matter) biomass. Unfortunately, the preparation of sequencing libraries from termite gut has proven difficult.
To address this challenge, a group at the University of California, Irvine, leveraged our RNA-seq technology to develop a novel method for metatranscriptomic analysis of the termite gut (Marynowska et al. 2017). Researchers generated libraries from the guts of four separate termite species using our SMARTer Stranded RNA-Seq Kit.
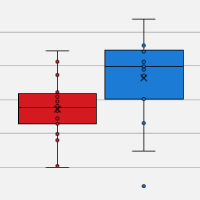
Following sequencing on an Illumina MiSeq® platform, analysis revealed that ~14% of all mRNA reads corresponded to carbohydrate metabolism and transport genes. Notably, researchers were able to use this data to identify 1,643 enzymes of interest spanning 116 families (see figure below).

Identification and fingerprinting of carbohydrate-active enzymes in termites. Metatranscriptomic analysis of the gut microbiome from each of four termite species reveals a unique "fingerprint" of carbohydrate-active enzymes dispersed across several different classes.
Why is this important?
Lignocellulosic biomass is the most plentiful source of raw material for producing biofuels, but current processing methods remain cost-prohibitive. Identifying enzymes that allow high-efficiency catalysis of biofuels would provide a cheap, renewable source of energy. RNA-seq approaches to identifying novel enzymes can be challenging due to the complexity of the termite gut microbiome and the low amount of starting material obtained—but our SMARTer Stranded RNA-Seq Kit rose to the challenge.
The future of the small
Recent technological advances in large-scale DNA and RNA sequencing methods have enabled us to perform metagenomics and metatranscriptomics with unprecedented resolution, but there are still difficulties. High sample complexity, low sample input, and large sample numbers all represent significant challenges to overcome, but our ThruPLEX DNA-seq and SMARTer Stranded RNA-seq technologies offer solutions to help researchers get the data they need, even from extremely low input amounts.
References
Marynowska, M. et al. Optimization of a metatranscriptomic approach to study the lignocellulolytic potential of the higher termite gut microbiome. BMC Genomics 18, 681 (2017).
Piddock, L. J. V. Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4, 629–636 (2006).
Product line overview
Browse our broad NGS portfolio, which provides unmatched sensitivity for all of your demanding sequencing applications.
RNA-seq learning center
Learn more about our RNA-seq technology and applications.
Tips for BCR/TCR profiling
Get started with NGS-based immune profiling of B- and/or T-cell repertoires
Technical notes
Technical notes for a wide variety of NGS applications.
Technology and application overviews
Overviews of our NGS technologies and applications.
FAQs and tips
Helpful information for successful NGS experiments.
DNA-seq protocols
Currently featuring protocols for integrating ThruPLEX kits with leading target enrichment systems.
Bioinformatics resources
Browse our library of bioinformatics software resources, including kit and reagent compatibility, user manuals, software notices, troubleshooting tips, and videos.
Webinars
Prerecorded webinars covering a variety of NGS applications.
Posters
Posters featuring Takara Bio NGS methods and data.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.