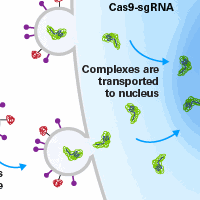
CRISPR/Cas9 gene editing is an RNA-programmable system which has democratized targeted genome modification by virtue of its simplicity and versatility. With this technology, gene editing is mediated by Cas9 nuclease and a single-guide RNA (sgRNA) which directs the Cas9 to a specific genomic locus. However, the utility of any genome modification system—for either basic research or therapeutic development—depends on its specificity (Sternberg and Doudna 2015). Early reports have warned of the frequent off-target effects of the CRISPR/Cas9 system (Fu et al. 2013; Hsu et al. 2013; Mali et al. 2013; Pattanayak et al. 2013), demonstrating the need for developing new methods that reduce these events.
The Guide-it CRISPR/Cas9 Gesicle Production System is a novel methodology that uses cell-derived nanovesicles, called gesicles, for co-delivery of active Cas9 protein complexed with a gene-specific sgRNA. Gesicles are loaded with Cas9-sgRNA ribonucleoprotein (RNP) complexes and can be added directly to target cells for CRISPR/Cas9 gene editing. Delivery of active Cas9 protein means no Cas9 coding gene is present in target cells, thus eliminating the problem of persistent and elevated Cas9 levels, which is common to plasmid-based delivery methods.
Tech Note
Reduced off-target effects with delivery of active Cas9 protein complexed with sgRNA using gesicle technology
Guide-it CRISPR/Cas9 Gesicle Production System
Introduction
Results
CRISPR/Cas9 gesicles reduce off-target effects compared to plasmid transfection
In a side-by-side comparison of Cas9-sgRNA delivery methods, the EMX1 gene was edited in HEK 293T cells either by treatment with gesicles loaded with Cas9-sgRNA RNP complexes or by transfection with plasmids encoding Cas9 and a gene-specific sgRNA (Figure 1). Following CRISPR/Cas9 modification, the EMX1 locus and a potential off-target locus (off-target 4) were PCR-amplified from crude cell extracts. The presence of indels was detected using Guide-it Resolvase (a mismatch-specific nuclease included in the Guide-it Mutation Detection Kit), followed by agarose gel electrophoresis. Densitometry (Cong et al. 2013) showed roughly equivalent indel formation at the EMX1 target locus between the two methods. As expected, plasmid transfection resulted in significant indel formation at the off-target 4 locus, while gesicle delivery resulted in no observable indel formation at this locus (Figure 1).

Figure 1. Reduced off-target effects with CRISPR/Cas9 gesicles. HEK 293T cells were either treated with gesicles loaded with Cas9-sgRNA RNP complexes or transfected with plasmids encoding Cas9 and an sgRNA against EMX1. After 72 hr, the EMX1 gene and a potential off-target locus (off-target 4) were amplified from the treated cells by direct PCR. Using the Guide-it Mutation Detection Kit, the amplicons were melted and rehybridized, and mismatched targets were cleaved using Guide-it Resolvase. A control sample that lacked Guide-it Resolvase was included for comparison (Control). The percentage of indel formation was determined by densitometry. No off-target effects were detected following the gesicle treatment.
To confirm the results of the resolvase assay, PCR amplicons of both the EMX1 locus and off-target 4 were subcloned and sequenced. Corroborating the previous results (Figure 1, above), plasmid delivery resulted in indel formation at both the EMX1 and off-target 4 loci (Figure 2). Gesicle delivery resulted in indel formation only at the EMX1 locus.

Figure 2. Sanger sequencing confirmed reduced off-target effects with CRISPR/Cas9 gesicles. EMX1 and off-target 4 PCR amplicons were subcloned using the Guide-it Indel Identification Kit. Sequencing data for the different clones were aligned with the wild-type sequence (underlined), revealing a range of deletions and insertions (highlighted in red) in both the EMX1 and off-target 4 sites when cells were treated with plasmid transfection. For the cells treated with gesicles, indels were detected only at the EMX1 target site, not at the off-target 4 site.
Conclusions
The Guide-it CRISPR/Cas9 Gesicle Production System is a novel methodology for the delivery of active Cas9-sgRNA RNP complexes to target cells for CRISPR/Cas9 gene editing. Delivery of these active RNP complexes in this manner prevents both the overexpression and genomic integration of Cas9 inherent to plasmid-based delivery. Therefore, Guide-it CRISPR/Cas9 Gesicles advance genome modification by enabling efficient editing of target loci while also reducing potential off-target effects.
Methods
Production of gesicles containing Cas9 protein and sgRNA
The target sgRNA against EMX1 was cloned into the prelinearized pGuide-it-sgRNA1 vector included in the Guide-it CRISPR/Cas9 Gesicle Production System. This cloned plasmid was added to the provided Guide-it Gesicle Packaging Mix. The Gesicle Packaging Mix contains lyophilized Xfect Transfection Reagent premixed with an optimized formulation of plasmids encoding Cas9 and all the other elements needed for gesicle production.
Modification of the EMX1 and off-target 4 genes in HEK 293T target cells
5.0 x 105 HEK 293T cells were plated in 24-well plates. 24 hr later, cells were either cotransfected with 500 ng each of plasmids encoding Cas9 and a sgRNA targeting EMX1 using Xfect Transfection Reagent, or treated with 30 µl of Cas9 gesicles (produced as described above).
Determination of indel formation
72 hr later after plasmid or gesicle treatment, the level of indel formation was determined using the Guide-it Mutation Detection Kit. Crude DNA extracts were prepared from cells. The modified EMX1 locus and potential off-target site (off-target 4) were amplified using direct PCR. The PCR amplicons were melted and rehybridized, then analyzed using the mismatch-specific nuclease, Guide-it Resolvase. The cleavage reactions were run on an agarose gel, and the percentage of DNA cleavage was determined by densitometry.
Sanger sequencing for confirmation of indels
EMX1 and off-target 4 PCR amplicons were subcloned using the Guide-it Indel Identification Kit and submitted for Sanger sequencing.
References
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–23 (2013).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–6 (2013).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–8 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839-43 (2013).
Sternberg, S. H. & Doudna, J. A. Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol. Cell 58, 568–574 (2015).
Related Products
CRISPR/Cas9 information
Choosing sgRNA design tools
Browse a collection of sgRNA design tools for Cas9-based genome editing experiments.
Choosing a target sequence for CRISPR/Cas9 gene editing
Learn how to design sgRNA sequences for successful gene editing.
The CRISPR/Cas9 system for targeted genome editing
Overview of CRISPR/Cas9 system for genome editing.
CRISPR/Cas9 genome editing tools
An overview of tools available for each step in a successful genome editing workflow.
Gene editing technical notes
Delivery of Cas9 and sgRNA to mammalian cells using a variety of innovative tools.
SNP engineering application note
Learn about a simple assay for sensitive detection of single-nucleotide substitutions in bulk-edited or clonal cell populations.
CRISPR/Cas9 gesicles overview
Learn about Guide-it CRISPR/Cas9 Gesicle Production System components and workflow.
CRISPR library screening webinar
Watch this webinar to learn how you can perform genome-wide lentiviral sgRNA screens easily.
Choosing an HDR template format
Watch a webinar on how to choose the right HDR template for knockin experiments.
Guide-it SNP Screening Kit FAQs
Get answers to frequently asked questions and view a video explaining the enzymatic assay.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.