Homology-directed repair FAQs
A powerful application of CRISPR/Cas genome editing technology involves the precise insertion or substitution of DNA sequences at specific genomic loci via the homology-directed repair (HDR) pathway. While this approach for engineering site-specific knockins is becoming increasingly popular, there are many ways to go about it, and its efficiency can vary widely depending on factors such as the target organism or cell type, the genomic locus, or the DNA template.
To maximize your likelihood for success with genome editing using HDR, our R&D team has compiled responses for the following frequently asked questions:
How do I design my HDR template?
The sequence of the HDR template should match the intended sequence of the targeted genomic locus, including the desired edit flanked by 5' and 3' ends that share homology with the wild-type genomic locus. (These 5' and 3' sequences are referred to as "homology arms.")
Sequences corresponding to desired edits (insertions, substitutions, etc.) should be positioned in the middle of the HDR template. The exact sequence of the HDR template is independent of the position of the single guide RNA (sgRNA) or the cut site of the Cas9 nuclease, but efficient editing requires a template design in which the sequence(s) to be inserted are in relatively close proximity to the cut site of the Cas9-sgRNA ribonucleoprotein (RNP). (See FAQ below for more detail.)
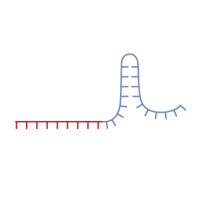
The figure below demonstrates the design of HDR templates for engineering a single nucleotide substitution or a longer insertion, respectively, at corresponding genomic loci.

Figure 1. Examples of HDR template designs for engineering a single nucleotide substitution or insertion of a sequence. In either case, the HDR template should encode the final desired edit with 5' and 3' flanking sequences homologous to the wild-type target site. Panel A. Design of an HDR template for engineering the mutation g.1488C>T in the RECQL4 gene. The mutated base is shown in red. Panel B. Design of an HDR template for inserting a myc tag at the C terminus of the UGT1A9 gene. In this case, three different sgRNAs were designed to target sites in close proximity to the location of the intended insertion.
What are the advantages and disadvantages of using single-stranded vs. double-stranded HDR templates for engineering gene knockins?
It has been demonstrated that the use of ssDNA results in lower toxicity and reduced frequencies of random integration relative to dsDNA (Roth et al. 2018; Li et al. 2017), benefits which can be especially important when working with difficult-to-engineer cell lines. For example, if you are seeking to fuse a fluorescent protein to an endogenous gene expressed in your target cells, the percentage of fluorescent cells in the overall edited population could be used as a proxy for the frequency of successful HDR. However, some subset of these fluorescent cells could have resulted from random integration of the HDR template in frame with a gene other than the intended genomic target. (Panel 4B in this poster includes data from our R&D team demonstrating this phenomenon.) Such events are less probable when ssDNA is used as a template for HDR.
References
Li, H et al., Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv doi: https://doi.org/10.1101/178905 (2017).
Roth, T.L et al., Reprogramming human T cell function and specificity with non-viral genome targeting. Nat. Lett. 559, 405–409 (2018).
How do ssODNs differ from ssDNA? When is it better to use one vs. the other?
Single-stranded oligo deoxynucleotides (ssODNs) are produced routinely via chemical synthesis and can be used for engineering smaller edits (e.g., single nucleotide substitutions or shorter insertions <50 nt) as HDR templates with a total maximum length of 200 nt. For engineering larger insertions that necessitate HDR templates >500 nt, we recommend the use of ssDNA that can be produced with the Guide-it Long ssDNA Production System.
With ssODN or ssDNA, is it advantageous to use HDR templates that are homologous to the sense or antisense strand at the genomic target region?
Currently, there is no clear rule regarding the preferential use of sense or antisense ssDNA for HDR templates longer than 200 nt. However, in the case of shorter single-stranded oligodeoxynucleotide (ssODN) templates, template polarity has been found to have an effect on efficiency (Bollen et al. 2018).
References
Bollen, Y., Post, J., Koo, B. & Snippert, H.J.G How to create state-of-the-art genetic model systems: strategies for optimal CRISPR-mediated genome editing. Nuc. Acid. Res. 46, 6435–6454 (2018).
What are optimal lengths for HDR template homology arms?
It has been demonstrated that there is an exponential relationship between homology arm length and knockin efficiency, with ssDNA templates including homology arms 350–700 nt in length providing optimal performance (Li et al. 2017). In our own studies involving hiPSCs, optimal performance was observed with 350-nt homology arms, and the use of longer arms did not substantially increase knockin efficiencies.
It is important to note that using longer homology arms will increase the molecular weight of your HDR template, and since the amounts of HDR template used for electroporation are usually measured in µg, longer homology arms will correspond with fewer mols (copies) of template being introduced in the target cells, possibly impacting the overall percentage of HDR.
References
Li, H et al., Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv doi: https://doi.org/10.1101/178905 (2017).
Is it helpful to use HDR templates that introduce silent mutations in the PAM sequence or sgRNA seeding region?
Cas9-sgRNA RNPs can re-cut a given genomic target following HDR if the editing outcome does not involve alteration of the PAM or sgRNA seeding region in the protospacer sequence, such that the RNP complex can still bind to the genomic target. Given this, a common strategy for increasing the percentage of successful HDR in coding regions is the incorporation of silent mutations in HDR templates that disrupt Cas9-sgRNA RNP binding without altering corresponding amino acid sequences.
If your PAM sequence is in a coding region and it can be silently mutated, it is advisable to do so to maximize the precision of genome editing and prevent subsequent Cas9 re-cutting and indel creation following HDR (Paquet et al. 2016). In seeking to mutate the PAM to disrupt RNP binding, please take into account other canonical PAM sequences (different from NGG) that can also be recognized less effectively by SpCas9 (i.e., NAG or NGA) (Zhang et al. 2014; Hsu et al. 2013) or Cas12a (i.e., TCTA, TCCA, or CCCA) (Yamano et al. 2017). Presented in the figure below are data demonstrating improved editing performance via incorporation of PAM mutations in HDR templates.
As mentioned above, the protospacer sequence can also be mutated. However, since some guide RNAs may tolerate a mismatch in the binding sequence, we recommend changing several bases as close to the PAM as possible in the seeding region of the sgRNA (Paquet et al. 2016; Kwart et al. 2017).

Figure 2. Improved genome editing performance via mutation of PAM sequences. The UGT1A9 gene was tagged with a myc tag (depicted in yellow, lowercase nucleotides) at its C terminus, just before the STOP codon (TGA, depicted in grey). Three different sgRNAs were designed to target genomic sites close to the intended insertion site, and all three of them were tested independently. ChiPSC18 cells were electroporated with RNP complexes together with two different HDR donors: one encoding for the intended myc-tag fusion with no additional mutations, and another encoding for the fusion as well as mutations in all three PAM sites of the corresponding sgRNAs. After electroporation, HDR efficiencies for the different samples were analyzed using the Guide-it SNP Screening Kit, which detects seamless insertion of the tag at its 5' and 3' ends, yielding green and red fluorescent signals, respectively. The highest signal for both wavelengths corresponded to the use of sgRNA3 in tandem with the HDR template with the mutated PAM sites. In parallel, HDR efficiencies were also analyzed using an RFLP-based method involving a new NdeI site created by the insertion of the myc tag. For this analysis, a signal could only be detected for cells edited using sgRNA3 in tandem with the HDR template carrying mutated PAM sites (i.e., the same cells that yielded the strongest fluorescent signals in the Guide-it screening assay).
References
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–32 (2013).
Kwart, D., Paquet, D., Teo, S. & Tessier-Lavigne, M. Precise and efficient scarless genome editing in stem cells using CORRECT. Nat. Protoc. 12, 329–354 (2017).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Yamano, T. et al. Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1. Mol. Cell 67, 633–645.e3 (2017).
Zhang, Y. et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 4, doi: 10.1038/srep05405 (2014).
Does the HDR insertion site need to be directly next to the PAM?
It is highly recommended that the HDR insertion site is positioned within 10 nt upstream or downstream of the Cas9-sgRNA RNP cut site. For wild-type SpCas9, this is located 3–4 nucleotides upstream of the PAM. It has been demonstrated that there is an inverse relationship between HDR efficiency and the distance between the insertion site and Cas9-gRNA RNP cut site (Paquet et al. 2016).
References
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
What are the effects of chromatin configuration on HDR efficiency?
It has been demonstrated that nucleosomes inhibit target cleavage by SpCas9 in vivo, suggesting that Cas9 activity is influenced by chromatin structure in addition to the inherent activity of a given sgRNA (Yarrington et al. 2018). Although DNA sequence is a significant determinant of site-specific indel profiles, it has been shown that packaging of DNA into chromatin may influence editing efficiency and the relative frequency of indels at a given locus (Chakrabarti et al. 2018). On the other hand, using imprinted genes as a test case, outcomes of gene editing events (i.e., the balance between HDR vs. NHEJ) were shown to be independent of chromatin state (Kallimasioti-Pazi et al. 2018).
References
Chakrabarti, A. M. et al. Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol. Cell 73, 1–15 (2018).
Kallimasioti-Pazi, E. M. et al. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLOS Biol. 16, e2005595 (2018).
Yarrington, R. M., Verma, S., Schwartz, S., Trautman, J. K. & Carroll, D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl. Acad. Sci. 115, 9351–9358 (2018).
What are additional methods for improving HDR efficiency?
The development of novel methods for improving HDR efficiency is currently a major focus area in the genome editing field. Toward this end, the following approaches have shown promise:
- Fusion or attachment of Cas9-sgRNA RNP and template (Aird et al. 2018; Savic et al. 2018; Carlson-Stevermer et al. 2017)
- Restriction of Cas9 expression to S and G2 phases of the cell cycle, when the propensity for HDR is highest (Gutschner et al. 2016)
- Development of base editing for engineering single nucleotide substitutions (Rees and Liu 2018)
- Application of various chemical compounds that increase the percentage of HDR (Wienert et al. 2018; Riesenberg and Maricic 2018)
References
Aird, E. J., Lovendahl, K. N., St. Martin, A., Harris, R. S. & Gordon, W. R. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 1, 54 (2018).
Carlson-Stevermer, J. et al. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 8, 1711 (2017).
Gutschner, T., Haemmerle, M., Genovese, G., Draetta, G. F. & Chin, L. Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep. 14, 1555–1566 (2016).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Riesenberg, S. & Maricic, T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 9, 2164 (2018).
Savic, N. et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 7, (2018).
Wienert, B. et al. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. bioRxiv 500462 (2018). doi:10.1101/500462
How can I increase the frequencies of edited clones with only one modified allele and the other one encoding the wild-type sequence?
One of the most common genome editing outcomes is the introduction of an SNP in one allele and an indel in the other, such that it is especially challenging to obtain clones in which only one allele is modified (encoding for the desired edit) and the other one is unedited. To more efficiently obtain clones with SNP/wild-type heterozygosity, researchers have developed two strategies (Paquet et al. 2016):
- Use an sgRNA with a cut site farther away from the insertion site (>10 nt)
- Use an equal mix of mutant and wild-type HDR templates (one encoding for the SNP, the other encoding for the wild-type allele, both with the PAM sequence modified as explained in the previous FAQ)
Reference
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
CRISPR/Cas9 information
Choosing sgRNA design tools
Browse a collection of sgRNA design tools for Cas9-based genome editing experiments.
Choosing a target sequence for CRISPR/Cas9 gene editing
Learn how to design sgRNA sequences for successful gene editing.
The CRISPR/Cas9 system for targeted genome editing
Overview of CRISPR/Cas9 system for genome editing.
CRISPR/Cas9 genome editing tools
An overview of tools available for each step in a successful genome editing workflow.
Gene editing technical notes
Delivery of Cas9 and sgRNA to mammalian cells using a variety of innovative tools.
SNP engineering application note
Learn about a simple assay for sensitive detection of single-nucleotide substitutions in bulk-edited or clonal cell populations.
CRISPR/Cas9 gesicles overview
Learn about Guide-it CRISPR/Cas9 Gesicle Production System components and workflow.
CRISPR library screening webinar
Watch this webinar to learn how you can perform genome-wide lentiviral sgRNA screens easily.
Choosing an HDR template format
Watch a webinar on how to choose the right HDR template for knockin experiments.
Guide-it SNP Screening Kit FAQs
Get answers to frequently asked questions and view a video explaining the enzymatic assay.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.