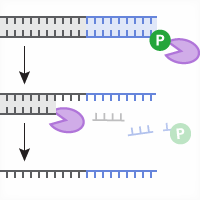
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system is the newest tool for targeted genome editing. This genome editing technique is the most flexible method so far. This system is based on two key components that form a complex: the Cas9 endonuclease and a target-specific RNA (single guide RNA or sgRNA) that guides Cas9 to the genomic DNA target site (Figure 1).
Double strand breaks that are introduced to genomic DNA by Cas9 nuclease can be repaired via the homologous recombination (HR) or nonhomologous end joining (NHEJ) cellular repair mechanisms. HR requires an identical (or nearly identical) DNA template to repair the break, and results in full correction of the DNA cleavage. However, DNA damage is more commonly repaired by NHEJ. In this case, cellular machinery ligates the two broken ends of the DNA without a template donor. This process is more error prone than HR, and often insertions and deletions (indels) are incorporated, resulting in a DNA sequence that differs from the wild-type sequence.
Learn more about CRISPR/Cas9 gene disruption »
Tech Note
Characterization of CRISPR/Cas9-introduced mutations using the Guide-it Indel Identification Kit
- A streamlined method for identifying indels introduced by CRISPR/Cas9 genome editing in a cell population
- Knockout of AcGFP1 using CRISPR/Cas9 genome editing
- Disruption of CD81 using CRISPR/Cas9 genome editing
The Guide-it Indel Identification Kit contains all components needed to amplify directly from cells, clone, and prepare modified target sites for DNA sequence analysis—in only 15 minutes.
Introduction
Results
A streamlined method for indel identification
Any cell in a population has the ability to correct endonuclease cleavage by either HR or NHEJ, resulting in a variety of DNA sequences at the target site in different cells. To identify the range of indels at the genomic DNA target site in a cell population after editing, we developed a streamlined workflow to amplify, clone, and analyze target sites by DNA sequencing (Figure 1).
This protocol uses a small sample of the cell population as a template for direct PCR amplification of the genomic region containing the endonuclease target site. The use of Terra PCR Direct Polymerase allows amplification directly from crude cell lysates without the need for genomic DNA purification (Figure 1, step 1). The resulting PCR pool of DNA fragments, containing different mutations at the target site, are cloned into a pUC19 vector using In-Fusion Cloning, a ligation-free cloning system (Figure 1, step 2). After introduction into optimized Stellar competent cells, colony PCR is used to amplify the inserts (Figure 1, step 3). Finally, DNA sequencing of the amplified inserts is used to identify indels (Figure 1, step 4).

Figure 1. Workflow for the Guide-it Indel Identification Kit
Knockout of AcGFP1 using CRISPR/Cas9 genome editing
CRISPR/Cas9 genome editing technology was applied to disrupt a single copy of AcGFP1 integrated in the genome of HT1080 cells (HT1080-AcGFP1). These cells exhibit strong green fluorescence (Figure 2, red trace). Therefore, AcGFP1 knockout can be assessed by monitoring GFP expression by flow cytometry. In cells that were co-transfected with two plasmids, one expressing Cas9 and the other expressing an sgRNA targeting AcGFP1, a significant percentage of the population (67%) lost GFP expression, due to mutation of the AcGFP1-encoding gene (Figure 2).

Figure 2. Flow cytometry analysis of HT1080-AcGFP1 cells. Cells were analyzed before (red line) and after (green) transfection with plasmids encoding Cas9 and an AcGFP1-targeting sgRNA.
The different indels created at the AcGFP1 locus in the cell population were identified using the Guide-it Indel Identification Kit (according to the protocol in Figure 1). Cells (2 x 105) from the transfected cell population were collected and used directly as a template for PCR amplification of the genomic AcGFP1-encoding gene targeted by Cas9/sgRNA. The amplified genomic DNA fragments (Figure 3, lane 1) contain a mixture of the different indels that occurred via NHEJ repair at the AcGFP1 locus following double strand breakage.
The PCR products were cloned into a pre-linearized pUC19 vector using In-Fusion Cloning. Eight of the resulting bacterial clones were selected for colony PCR using Terra PCR Direct Polymerase with primers that amplified the insert. The amplified DNA was analyzed on an agarose gel, and all colonies showed a band of the correct size (Figure 4).

Figure 3. PCR product amplified from the Cas9 sgRNA target sequence.

Figure 4. Agarose gel of colony PCR products from eight clones. Lane M: Marker; Lane 1-8: Individual clones.
The PCR products were sequenced using the provided sequencing primers. The obtained DNA sequence was aligned with the wild-type AcGFP1 sequence (Figure 4). The sequences showed a high rate of an inserted "T" at the Cas9/sgRNA cleavage site in the different clones. One clone had a four nucleotide deletion immediately upstream of the PAM site. Only one clone contained the wild-type AcGFP1 sequence (Figure 5).

Figure 5. Sequencing results of the AcGFP1 target sequences obtained from eight different clones.
Disruption of CD81 using CRISPR/Cas9 genome editing
CD81 is a membrane receptor expressed in most cell types. Using a FITC-tagged antibody against CD81, expression can be monitored by flow cytometry (Figure 6).

Figure 6. Flow cytometry analysis of HeLa cells, labeled with and without an anti-CD81-FITC antibody.
The CD81 gene was targeted for CRISPR/Cas9 genome editing by transfecting HeLa cells with two plasmids encoding Cas9 and a CD81-targeting sgRNA, respectively. Five days after transfection the cells were harvested, labeled with an anti-CD81-FITC antibody, and analyzed by flow cytometry (Figure 7). The analysis indicated that CD81 expression was lost in approximately 51% of the cell population.

Figure 7. Flow cytometry analysis of HeLa cells labeled with the anti-CD81-FITC antibody after transfection with Cas9 and sgRNA targeting the CD81 gene.
The Guide-it Indel Identification Kit was used to characterize the nature of the indels created by Cas9/sgRNA in the cell population. Cells from the same population analyzed in Figure 6, which contained CD81 expressing and non-expressing cells, were used as template for direct PCR amplification of the targeted CD81 genomic site (Figure 8).
After cloning the PCR products, six bacterial clones were picked for colony PCR amplification of the CD81 target sequence using Terra PCR Direct Polymerase. The amplified DNA was analyzed on an agarose gel, and all colonies showed a band of the correct size (Figure 9).

Figure 8. PCR products from amplification of the targeted CD81 site from the cell population analyzed in Figure 6. Lane M: Marker; Lane 1: Amplified CD81 target site (* indicates location of PCR product).

Figure 9. Agarose gel of colony PCR products directly from six clones. Lane M: Marker; Lane 1-6: Individual clones.
The amplified PCR products were sequenced using the provided sequencing primer, and the sequencing data were aligned to the wild-type CD81 sequence (Figure 10). The obtained sequencing data from the different clones showed a wide variety of deletions and insertions at the targeted CD81 gene, emphasizing the variation in the outcome of NHEJ repair between individual cells.

Figure 10. Sequence analysis of six clones obtained from cells transfected with Cas9/sgRNA targeting the CD81 gene.
Related Products
Gene editing resources
Product finder
Use this gene editing product finder to quickly locate kits for screening, delivery, and downstream assays.
CRISPR tools and information
What is CRISPR/Cas9? Need help designing the best guide RNAs? Learn all this and more.
Genome-wide library screening
Learn how to conduct a CRISPR/Cas9 guide RNA library phenotypic screen and view data demonstrating the use of our library.
CRISPR/Cas9 knockouts
Technotes and tools used to create or study CRISPR-Cas9-mediated gene knockouts (indels).
CRISPR/Cas9 knockins
Information, technotes and tools used to create or study gene knockouts by CRISPR/Cas9 and HDR.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.