SMART-Seq Mouse TCR (with UMIs)
SMART-Seq Mouse TCR (with UMIs), powered by SMART (Switching Mechanism at 5’ end of RNA Template) technology, enables detection of mouse TCR sequences with sensitivity and no bias. The kit combines NGS with a 5’-RACE approach to capture the full-length V(D)J variable regions of mouse TRA and TRB genes.
SMART-Seq Mouse TCR (with UMIs), powered by SMART (Switching Mechanism at 5’ end of RNA Template) technology, enables detection of mouse TCR sequences with sensitivity and no bias. The kit combines NGS with a 5’-RACE approach to capture the full-length V(D)J variable regions of mouse TRA and TRB genes.
The kit is designed to work with a range of RNA input amounts (RIN >7) and has been shown to generate high-quality sequencing libraries from as little as 1 ng–1 µg of total RNA obtained from mouse spleen or T cells; 10 ng–1 µg of total RNA obtained from mouse whole blood, bone marrow, or thymus; or from 100 to 10,000 purified mouse T cells. Libraries can be generated to obtain both alpha- and beta-chain diversity information. This profiling kit also includes unique molecular identifiers (UMI)—making it possible to remove reads derived from PCR duplicates and sequencing errors—thus ensuring more accurate and reliable results. Generate up to 384 multiplexed Illumina libraries using the Unique Dual Index kits (Cat. Nos. 634752–634756; sold separately).
Major benefits of this kit:
- Compatibility with high-quality RNA isolated from a variety of sample types
- Ability to incorporate UMIs to correct for PCR and sequencing errors
- Cost savings on sequencing with flexibility to sequence full-length V(D)J or CDR3 regions only
- Cost savings by pooling up to 384 TCR libraries together with UDIs
Overview
-
- Compatible with a wide range of sample inputs—1 ng–1 µg of total RNA from mouse spleen or T cells; 10 ng–1 µg of total RNA obtained from mouse whole blood, bone marrow, or thymus; or from 100–10,000 purified mouse T cells
- Sensitivity meets simplicity—sensitive detection of TCR clonotypes with a single primer pair for each TCR (alpha or beta) subunit per reaction
- Sequencing flexibility—either full-length V(D)J or CDR3 only
- UDI implementation and UMI-based correction—decrease data errors in your NGS libraries
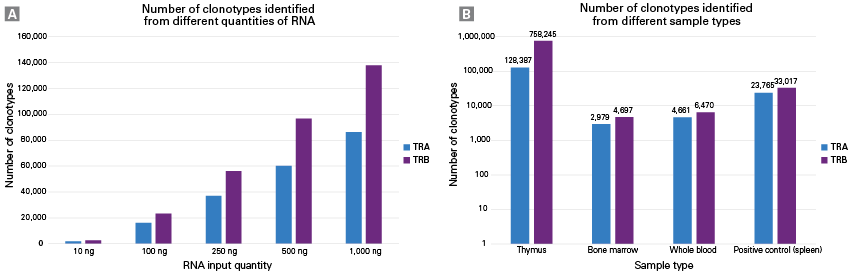
Evaluation of mTCRv2 kit performance across various sample types. Panel A. TCR libraries were made for both alpha (blue) and beta (purple) chains in 10, 100, 250, 500, and 1,000 ng of mouse spleen RNA with the mTCRv2 workflow. The resulting TCR libraries were sequenced on the Illumina NextSeq platform, generating 2 x 150 bp reads. Sequencing outputs were down sampled to ~15 million reads. Panel B. The mTCRv2 workflow was performed using 200 ng RNA isolated from four different sample types. The resulting TCR libraries were sequenced on NextSeq, generating 2 x 150 bp reads. Sequencing outputs were down sampled to 20 million reads for each sample type. Data was processed using Cogent NGS Immune Profiler software, and bar plot was generated to show clonotype counts for each individual input amount or sample type. TRA = TCR alpha chain, TRB = TCR beta chain.
More Information
Applications
Mouse TCR repertoire analysis (TCRα and TCRβ subunits) for studies such as:
- Analysis of pathogen-specific TCRs for identification of TCR biomarkers correlated with protection or amelioration of various infectious diseases
- Clonal organization and diversity of TCRs in tissues and circulation underlying migration, maintenance, and homeostasis of T lymphocytes
- Longitudinal analysis of immune aging dynamics and how changes in TCR repertoires affect responses to vaccines and other treatments in older adults
Additional product information
Please see the product's Certificate of Analysis for information about storage conditions, product components, and technical specifications. Please see the Kit Components List to determine kit components. Certificates of Analysis and Kit Components Lists are located under the Documents tab.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.