634352
SMART-Seq® Mouse BCR (with UMIs)
24 Rxns
USD $1406.00
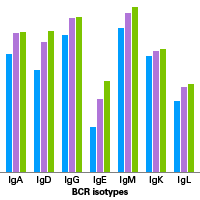
SMART-Seq Mouse BCR (with UMIs) enables users to analyze B-cell receptor (BCR) repertoires from 10 ng–1 µg high-integrity (RIN>7) bulk total RNA samples from spleen, bone marrow, or peripheral blood mononuclear cells (PBMCs) or RNA extracted from mouse whole blood (RNA input ≥100 ng) or lymph nodes. The kit can be used to generate data for both heavy (IgG, IgM, IgA, IgD, and IgE) and light chains (IgK, IgL) of mouse B-cell receptors.
Notice to purchaser
Our products are to be used for Research Use Only . They may not be used for any other purpose, including, but not limited to, use in humans, therapeutic or diagnostic use, or commercial use of any kind. Our products may not be transferred to third parties, resold, modified for resale, or used to manufacture commercial products or to provide a service to third parties without our prior written approval.
Cat. #
Product
Size
Price
License
Quantity
Details
634752
Unique Dual Index Kit (1–96)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (1–96) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634753
Unique Dual Index Kit (97–192)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (97–192) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634754
Unique Dual Index Kit (193–288)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (193–288) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634755
Unique Dual Index Kit (289–384)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (289–384) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634756
Unique Dual Index Kit (1–24)
24 Rxns
USD $237.00
The Unique Dual Index (UDI) Kit (1–24) contains 24 pairs of unique dual-indexed PCR primers, which are a subset of the Unique Dual Index Kit (1–96), Cat. No. 634752. They can be used to generate up to 24 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
744970.5
NucleoMag® NGS Clean-up and Size Select
5 mL
USD $119.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.5 consists of a bottle containing 5 ml of bead suspension.
744970.50
NucleoMag® NGS Clean-up and Size Select
50 mL
USD $805.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.50 consists of a bottle containing 50 ml of bead suspension.
744970.500
NucleoMag® NGS Clean-up and Size Select
500 mL
USD $5545.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.500 consists of a bottle containing 500 ml of bead suspension.
634353
SMART-Seq® Mouse BCR (with UMIs)
96 Rxns
USD $4983.00
SMART-Seq Mouse BCR (with UMIs) enables users to analyze B-cell receptor (BCR) repertoires from 10 ng–1 µg high-integrity (RIN>7) bulk total RNA samples from spleen, bone marrow, or peripheral blood mononuclear cells (PBMCs) or RNA extracted from mouse whole blood (RNA input ≥100 ng) or lymph nodes. The kit can be used to generate data for both heavy (IgG, IgM, IgA, IgD, and IgE) and light chains (IgK, IgL) of mouse B-cell receptors.
Notice to purchaser
Our products are to be used for Research Use Only . They may not be used for any other purpose, including, but not limited to, use in humans, therapeutic or diagnostic use, or commercial use of any kind. Our products may not be transferred to third parties, resold, modified for resale, or used to manufacture commercial products or to provide a service to third parties without our prior written approval.
Cat. #
Product
Size
Price
License
Quantity
Details
634752
Unique Dual Index Kit (1–96)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (1–96) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634753
Unique Dual Index Kit (97–192)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (97–192) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634754
Unique Dual Index Kit (193–288)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (193–288) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634755
Unique Dual Index Kit (289–384)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (289–384) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634756
Unique Dual Index Kit (1–24)
24 Rxns
USD $237.00
The Unique Dual Index (UDI) Kit (1–24) contains 24 pairs of unique dual-indexed PCR primers, which are a subset of the Unique Dual Index Kit (1–96), Cat. No. 634752. They can be used to generate up to 24 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
744970.5
NucleoMag® NGS Clean-up and Size Select
5 mL
USD $119.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.5 consists of a bottle containing 5 ml of bead suspension.
744970.50
NucleoMag® NGS Clean-up and Size Select
50 mL
USD $805.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.50 consists of a bottle containing 50 ml of bead suspension.
744970.500
NucleoMag® NGS Clean-up and Size Select
500 mL
USD $5545.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.500 consists of a bottle containing 500 ml of bead suspension.
634351
SMART-Seq® Mouse BCR (with UMIs)
4 x 96 Rxns
USD $18159.00
SMART-Seq Mouse BCR (with UMIs) enables users to analyze B-cell receptor (BCR) repertoires from 10 ng–1 µg high-integrity (RIN>7) bulk total RNA samples from spleen, bone marrow, or peripheral blood mononuclear cells (PBMCs) or RNA extracted from mouse whole blood (RNA input ≥100 ng) or lymph nodes. The kit can be used to generate data for both heavy (IgG, IgM, IgA, IgD, and IgE) and light chains (IgK, IgL) of mouse B-cell receptors.
Notice to purchaser
Our products are to be used for Research Use Only . They may not be used for any other purpose, including, but not limited to, use in humans, therapeutic or diagnostic use, or commercial use of any kind. Our products may not be transferred to third parties, resold, modified for resale, or used to manufacture commercial products or to provide a service to third parties without our prior written approval.
Cat. #
Product
Size
Price
License
Quantity
Details
634752
Unique Dual Index Kit (1–96)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (1–96) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634753
Unique Dual Index Kit (97–192)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (97–192) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634754
Unique Dual Index Kit (193–288)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (193–288) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634755
Unique Dual Index Kit (289–384)
96 Rxns
USD $623.00
The Unique Dual Index (UDI) Kit (289–384) contains 96 pairs of unique dual-indexed PCR primers that can be used to generate up to 96 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
634756
Unique Dual Index Kit (1–24)
24 Rxns
USD $237.00
The Unique Dual Index (UDI) Kit (1–24) contains 24 pairs of unique dual-indexed PCR primers, which are a subset of the Unique Dual Index Kit (1–96), Cat. No. 634752. They can be used to generate up to 24 Illumina-compatible sequencing libraries and are recommended with several of Takara Bio’s RNA-seq and DNA-seq kits.
744970.5
NucleoMag® NGS Clean-up and Size Select
5 mL
USD $119.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.5 consists of a bottle containing 5 ml of bead suspension.
744970.50
NucleoMag® NGS Clean-up and Size Select
50 mL
USD $805.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.50 consists of a bottle containing 50 ml of bead suspension.
744970.500
NucleoMag® NGS Clean-up and Size Select
500 mL
USD $5545.00
NucleoMag NGS Clean-up and Size Select employs scalable, automation-friendly magnetic bead technology to enable efficient cleanup of DNA or RNA fragments. The flexible protocol enables processing of 50–150 µl input volumes and allows for enrichment of fragments ranging in size from 150–800 bp. NucleoMag NGS Clean-up and Size Select consists of paramagnetic beads suspended in binding buffer that selectively bind DNA and RNA fragments based on the volume ratio of bead suspension to sample. After magnetic separation and removal of supernatant, the beads are washed with ethanol followed by a short drying step. Following elution in low-salt buffer or water, the highly purified DNA is free of contaminants such as nucleotides, primers, adapters, adapter dimers, enzymes, buffer additives, or salts, and can be used directly in downstream applications. NucleoMag NGS Clean-up and Size Select provides comparable performance and employs the same dilution ratios for size selection as magnetic bead-based solutions from other providers, such that it can be incorporated in existing workflows without the need for protocol adjustment or optimization.
Cat. # 744970.500 consists of a bottle containing 500 ml of bead suspension.