Seeker Spatial Transcriptomics Kit resources
Accelerate discoveries with high-resolution, whole-transcriptome expression maps
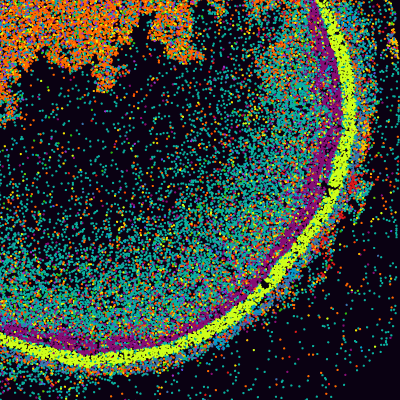
The Seeker tile preserves spatial information of the tissue transcriptome by capturing RNA on a monolayer of tightly-packed, spatially-indexed, 10 μm beads.
- Map the whole transcriptome—perform analysis that is not limited by predetermined targets or throughput, enabling true unbiased discovery
- Use your current lab instrumentation—leverage standard molecular biology techniques and instrumentation; no new specialized instrumentation or microscopy expertise needed
- Investigate samples from a wide array of organisms and tissues—analyze any polyadenylated RNA
What our customers are saying
Seeker doesn’t need a large upfront investment in equipment and lab automation, the perfect solution for starting with spatial transcriptomics. The protocol is straightforward and doesn’t require any specific and lengthy training. The possibility of sampling genes in an unbiased way and without having to design a panel makes Seeker the perfect technology for discovery experiments.
Simone Picelli, PhD
Team Leader, Single-Cell Genomics Platform
Institute of Molecular and Clinical Ophthalmology Basel
In our field of research dealing with rare T-cell subsets in immunological disease and cancer, unbiased characterization of cellular spatial context is an invaluable tool towards understanding cell niche composition, cell-to-cell interactions and disease pathogenesis. However, these approaches have thus far been hampered by poor resolution, which obfuscates underlaying biology. Compared to other competitors, the Seeker assay allows for cost-effective, near-cellular resolution profiling using a quick and simple protocol without compromises in data quality or sensitivity. These qualities empower researchers of all backgrounds to jump into the spatial genomics field and profile their tissue of interest.
Kelvin Chen, PhD
Assistant Professor, IFReC
Osaka University
Workflow

Simple, single-day workflow plugs directly into existing sequencing workflows. Once your tissue section is placed on the Seeker tile, mRNA is captured and hybridized on the spatially-indexed beads before reverse transcription. The indexed beads are then dissociated from the tile and the tissue is digested, followed by second strand synthesis and cDNA amplification. Purified cDNA undergoes NGS library preparation and is sequenced. The resulting FASTQ files are processed by the Seeker Analysis Pipeline to create a continuous whole transcriptome spatial map of your tissue section.
Sized to meet your needs

The Seeker kit produces gene expression maps with sharp spatial boundaries using tiles available in two convenient sizes: 3 mm x 3 mm and 10 mm x 10 mm. The Seeker Analysis Pipeline mapped spatial gene expression within a mouse hippocampus section (top row) and whole mouse brain (bottom row). Expression patterns of the top five spatially variable genes are shown.
Seeker protocol videos
Watch how to produce sequencing-ready libraries with spatial information from fresh-frozen tissues.
Seeker FAQs
Learn more about using Seeker technology for single-cell spatial biology research.
Advancing discovery with spatial multiomics—accessible tools for high-resolution insights
Webinar series: Advancing discovery with spatial multiomics—accessible tools for high-resolution insights.
Takara Bio USA, Inc.
United States/Canada: +1.800.662.2566 • Asia Pacific: +1.650.919.7300 • Europe: +33.(0)1.3904.6880 • Japan: +81.(0)77.565.6999
FOR RESEARCH USE ONLY. NOT FOR USE IN DIAGNOSTIC PROCEDURES. © 2025 Takara Bio Inc. All Rights Reserved. All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at takarabio.com.